
eDNA degrades over time as it is exposed to UV radiation and other factors in the environment. Regular autonomous sampling enables managers to monitor rare and infrequent targets that would otherwise be missed by traditional sampling methods. Illustration: Fiona Martin © 2020 MBARI
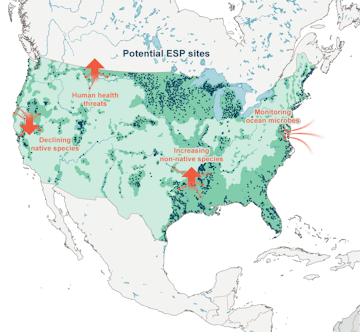
The ESP is a versatile and economical tool for monitoring key changes in aquatic health. This monitorial might include tracking the presence of native and non-native species, identifying harmful algal blooms, tracking pollution in our nation’s waterways, and studying ocean microbes. Illustration: Fiona Martin © 2020 MBARI
Monitoring eDNA at Scott Creek from MBARI on Vimeo.
Scott Creek, just north of Santa Cruz, California, is an optimal location for testing the application of eDNA monitoring in a freshwater stream. The creek is one of the southernmost habitats for endangered coho salmon and other species of interest like steelhead trout. The area is also at risk for the arrival of non-native New Zealand mudsnails and striped bass. NOAA has documented fish counts of coho salmon for decades at the site, providing a consistent historical dataset to compare against eDNA samples.


